|
Supervisor: Prof. Preeti Srivastava, DBEB IIT Delhi
March 2021- August 2022
Motivation: Temperature-sensitive (TS) plasmids are those plasmids that can replicate till a permissible temperature (say 30°C),
but they lose their ability to replicate at higher temperature (say 35°C), and thus, the plasmid is lost by the cell in successive replication cycles.
TS plasmids are important in synthetic biology as they give the ability to control the burden on any cell by altering the
plasmid replication as a function of temperature. They can be used in genetic engineering and manipulation as delivery vectors,
for inducible replication by supplying protein for transient expression and various other features. Once the plasmid has done the work and has served the purpose,
it becomes a burden to the cell, and thus, to reduce the burden, it is important to make the cell plasmid free. The TS plasmid can be cured by raising the temperature. |
|
|
Supervisor: Prof. Richard Murray, CDS Caltech
May 2020- August 2020
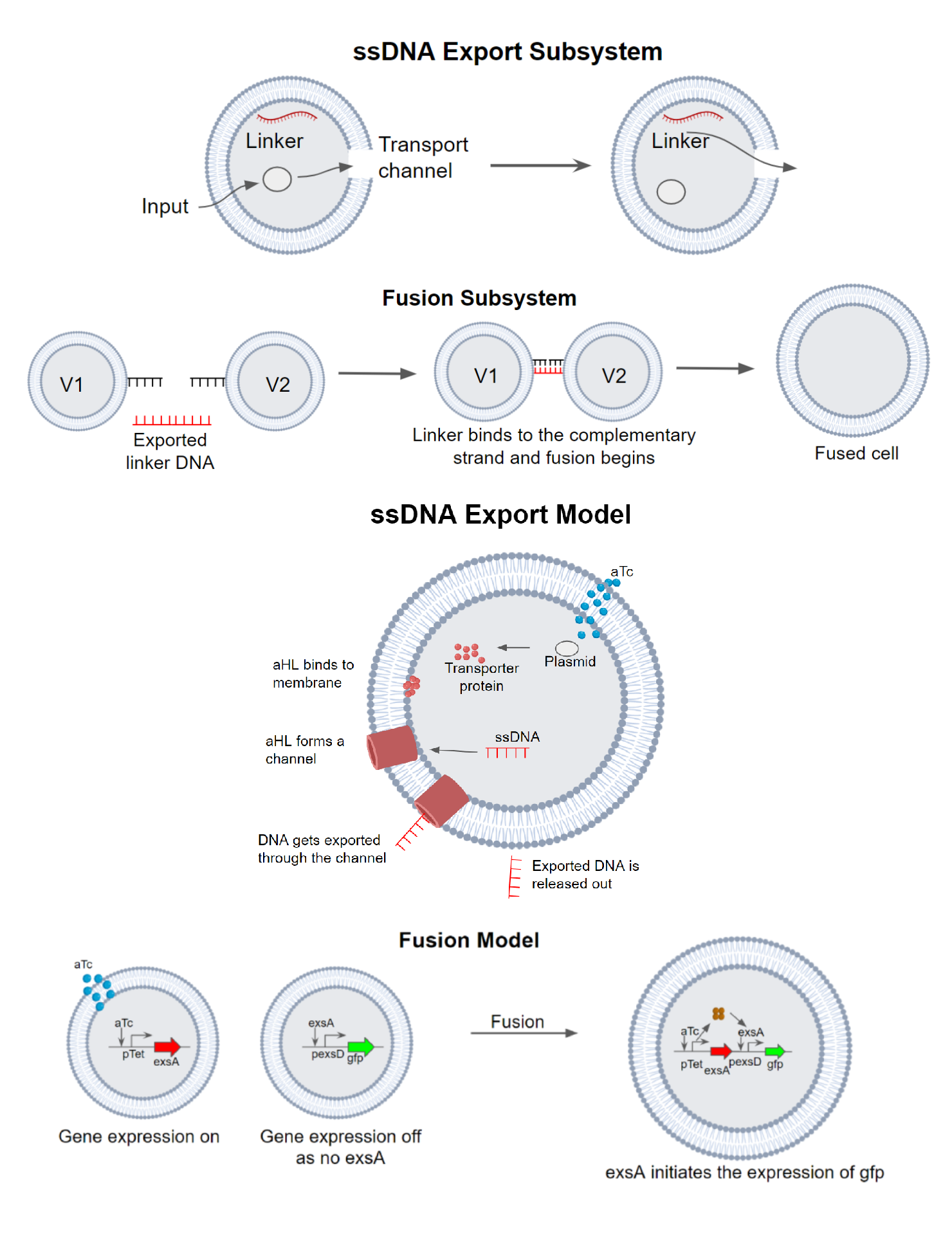
Motivation: Cell fusion is an important process for various cellular processes including cell-cell communication,
drug delivery, cell transfection and other therapeutic processes. For synthetic cells, designing and controlling spatial
and temporal biological reactions have been a challenging process. Targeted vesicle fusion has been shown as a promising
approach to control interactions between compartments selectively. Membrane fusion is also important to initiate protein
synthesis in synthetic cells. Thus, modeling and designing a biomolecular circuit is an important step.
|
|
|
Supervisor: Prof. Shaunak Sen, EE IIT Delhi
August 2019- December 2020
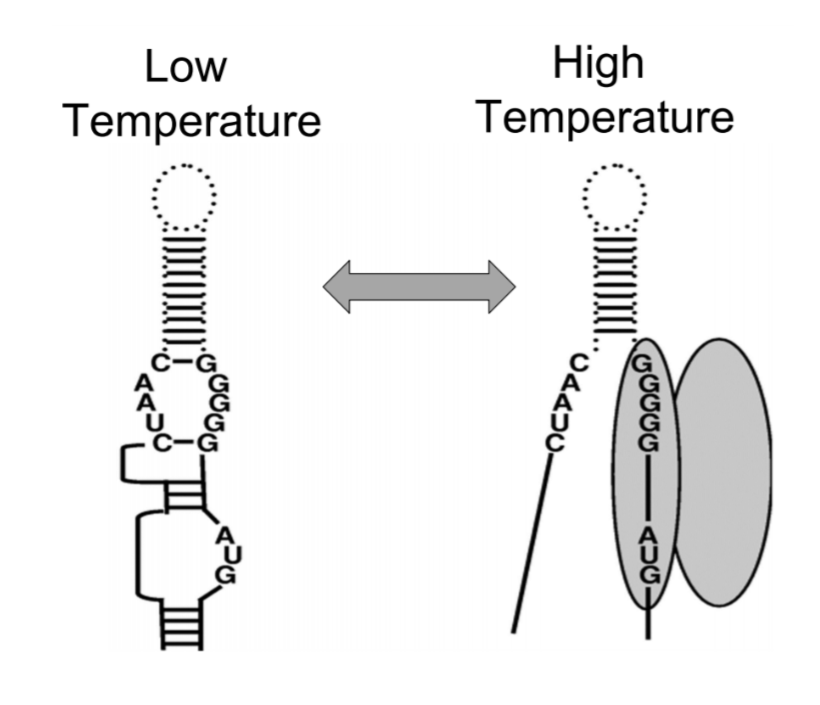
Motivation: RNA Thermometers (RNATs) are temperature-sensitive RNA molecules that can control protein expression.
RNATs alter their secondary structure in response to temperature fluctuations. They are present in the 5’
UTR region of mRNA sequence. As temperature increases, the hydrogen bonds break/ melt and thus, expose the
regions like ribosome binding sites that affect translation, and permit translation. This structural transition
can depend on either the heat shock or cold shock response resulting in either exposing or hiding of RBS.
Thus, it was important to understand the molecular mechanism or the structural transition that takes place by temperature change in some naturally occurring RNATs.
|
|
|
Supervisor: Prof. Preeti Srivastava, DBEB IIT Delhi
December 2021- October 2022
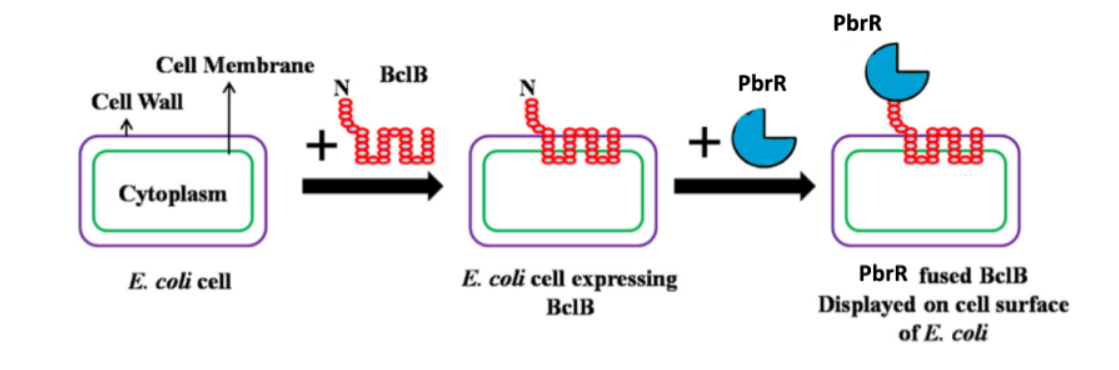
Motivation: Lead poisoning is a catastrophe affecting millions of people around the world.
Having a biosensor that could sense lead and also recover it from water could solve this major problem.
Previously developed whole-cell lead biosensors cater to a limited sample type i.e. soil samples.
Only a few studies have expanded the sample type to wastewater samples, which is one of the main
focuses of our biosensor. Whole-cell lead biosensors are intensity-based. Also, industries use various
physicochemical methods like chemical precipitation etc for lead removal from industrial wastewater,
which brings down the lead concentration from 200-500 mg/L to 0.4 mg/L, but is still ten-fold more
than the permissible limit and resulting in poor recovery efficiency beyond this concentration.
|
|